Our interests comprise basic and applied research, with three main areas:
- To understand the quality control and trafficking of transmembrane proteins (especially of major histocompatibility complex (MHC) class I molecules) between intracellular compartments and the cell surface;
- To understand and manipulate peptide selection by MHC class I molecules for basic research and practical application;
- To develop novel methods for analysis and manipulation of cells and with microparticles or engineered surfaces.
These areas often overlap, and we are especially interested in any synergies that develop from this interaction. We use methods from cell biology, biochemistry, biophysics, biotechnology, and computational biology.
MHC class I quality control and trafficking
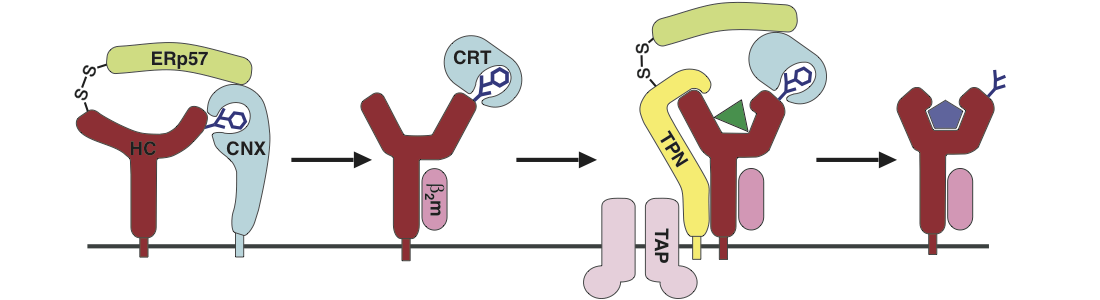
Our favourite model system are MHC class I molecules, extremely polymorphic proteins that are present on all nucleated cells. In humans, they are called HLA molecules. They play a central role in the mammalian immune defence against viruses, intracellular bacteria, and cancer, since they carry small peptides from intracellular proteins to the cell surface, where they can be surveyed by cytotoxic T lymphocytes (CTL) (a process called antigen presentation). If a specific CTL recognizes an unusual peptide being made inside the cell (e.g. originating from a viral or tumor protein), they induce the cell to undergo apoptosis. This way, the production sites of viruses are eliminated. MHC class I molecules are ideal model systems for protein folding, ligand binding, and intracellular transport, since several well-studied allotypes exist and many reagents to study them are available. They are at the core of the antiviral and antitumor response and are therefore in the focus of modern tumor immunotherapy.
Peptide binding to MHC class I molecules is a tightly controlled process. They do not just bind any peptide but only those that have the right length (8-10 amino acids) and sequence (some residues contact the binding site). Binding of such high-affinity peptides stabilizes the structure of the entire molecule, and trafficking of class I to and from the cell surface is dependent on the presence of the bound peptide ligand, i.e., class I molecules bound to low-affinity peptides or devoid of any peptide are usually kept inside the cell to avoid binding of extracellular peptides and thus the killing of a healthy cell.
In order to understand the mechanisms behind MHC class I quality control, our main questions are: What changes are brought about in the structure of the class I molecule by the peptide? How are these changes read out by the cellular quality control systems? How is the decision made to localize a class I molecule either inside the cell or on its surface? Why are different class I allotypes transported to the cell surface at different speeds?
MHC class I molecules bind peptides in the endoplasmic reticulum (ER) and then travel along the secretory pathway to the cell surface like any transmembrane protein. We have shown that trafficking of different class I allotypes is regulated by the ER chaperone matrix in a proteostatic fashion [Fritzsche et al., 2015]. Improperly loaded class I molecules are recognized for intracellular retention or retrieval to the ER [Garstka et al., 2007]. We also found that suboptimally loaded class I molecules are recognized by calreticulin in the cis-Golgi [Howe et al., 2009], and that this quality control system surveys the flexibility of the peptide binding site as an indicator for insufficient peptide binding and folding. By introducing an artificial disulfide bond, we stabilized the binding site, and those class I disulfide mutants show an increased in vivo stability at the cell surface even in a suboptimally loaded form [Hein et al., 2014].
At the end of their lifespan, class I molecules are endocytosed from the cell surface and destroyed in lysosomes. We have shown that loss of the MHC class I light chain, beta-2 microglobulin (β2m) prevents recycling from early endosomes and leads to the rapid destruction of the molecule [Montealegre et al., 2015]. We are gradually finding out how the “free” heavy chains are recognized inside the cell for routing to endosomes. This work is significant because class I endocytosis is known to affect the surface levels of other receptors.
Viruses often try to block peptide loading and trafficking of MHC class I molecules. Viral proteins that interfere with class I peptide presentation are called immunoevasins, and they are an interesting tool to further examine class I quality control mechanisms. We have shown that the gp40/m152 protein of the murine cytomegalovirus (mCMV) holds class I molecules in the ER-Golgi intermediate compartment by direct interaction [Janssen et al., 2016]. We continue to investigate the molecular mechanism of this interaction, and the retention of gp40 itself.
MHC class I peptide selection and its practical application
Peptide binding in the lumen of the ER is aided by the proteins of the so-called peptide loading complex, among them the chaperone tapasin, which imposes thermodynamic control on the peptide binding process and thus leads to stable peptide binding [Schneeweiss et al., 2009; Praveen et al., 2010]. We have shown with a variety of methods ranging from molecular dynamics simulations to fluorescence microscopy that binding and function of tapasin respond to the native unfolding of the class I peptide binding site; thus, some class I molecules require tapasin to a greater extent than others for peptide loading [Garstka et al., 2011; Abualrous et al., 2015].
Class I molecules select their peptides from a large intracellular pool, but they bind only a restricted set of peptides of certain length and sequence. The binding of such high-affinity peptides changes structure and flexibility of the class I peptide binding site and stabilizes the entire molecule. Due to their conformational instability, no crystal structure of empty, peptide-free class I molecules exist, but we have shown by molecular dynamics (MD) simulations that some parts of the peptide binding site are intrinsically mobile, i.e. natively unfolded, in the absence of peptide [Zacharias and Springer, 2004; Hein et al., 2014].
Occupation of one part of the binding site, the so called F-pocket, by a partial ligand is sufficient to quench the flexibility of the binding site [Saini et al., 2013; Abualrous et al., 2015]. These small molecules help class I molecules to fold and bind peptides, and they can also be used to trigger the exchange of a bound peptide for a peptide of choice; in this way, the dissociation of a bound peptide can be accelerated by a thousand fold [Saini et al., 2013; Saini et al., 2015]. The understanding of peptide binding opens the door to its manipulation: The usage of such small molecules facilitates T cell staining with MHC class I tetramers, and will thus improve tumor epitope discovery, diagnosis, and therapy.
Novel methods using microparticles and engineered surfaces
Our biotechnological projects develop novel techniques for research and application. Since 2007, we have used micrometer-sized polyelectrolyte capsules for various approaches and demonstrated, for instance, the introduction of hydrophilic solutes into mammalian cells by microencapsulation and controlled release [Palankar et al., 2009; Studer et al., 2010]. Just recently, we have developed a microcapsule-based sandwich assay for highly sensitive and robust protein and nucleic acid detection [Verma et al., 2016]. For this, the microcapsules are chemically coated with detector molecules such as antibodies that specifically bind an analyte of interest and are subsequently detected by flow cytometry. We were thus able to detect the blood cancer biomarker beta-2 microglobulin in a pico- to femtomolar concentration range. The assay allows rapid quantitative measurements, while providing high sensitivity and selectivity at very small sample quantities. We aim to further develop our microcapsules such that, in the future, they can be used as universal tools for the detection of a broad range of analytes.
Another biotechnical approach involves patterned surfaces to manipulate membrane proteins of live cells. They are an extremely useful tool to especially study ligand binding of cell surface receptors in their native environment. With antibodies reprinted on a glass surface, we were not only able to specifically array a distinct subpopulation of MHC class I molecules into a defined pattern on the surface of live cells but also to observe the specific binding of a fluorescently labeled index peptide by microscopy [Dirscherl et al., 2017]. The observation and measurement of ligand binding to captured cell surface receptors in defined conformations apply to many problems in cell biology and thus represent a promising tool in the field of biosensors.
Some articles that feature our work:
- Forscher der Jacobs University Bremen gründeten eine Firma für biomedizinische Reagenzien (2019)
- Discovery by Jacobs University researchers facilitates detection of immune cells that fight tumors and viruses (2015)
- Jacobs University Professor publishes joint study with Nobel laureate for Medicine (2014)
- Modern man inherited immune gene from Neanderthals (2013)
- Immune system monitoring improved (2013)
- New biotechnology cooperation between Jacobs and Hochschule Bremen is funded by BMBF with almost €1m (2012)
- Esther Ghanem, jeune boursière, raconte (about Esther Ghanem, a PhD student in our group, from the Lebanese newspaper L’Orient Le Jour, in French, 2011)
- Controlling the Immune Reaction (Jacobs, 2010)
- Micro shuttles and laser remote control (Jacobs, 2009)
- Light-switched drug delivery (MIT Technology Review, 2009)
- Transport mechanism for key immune proteins (Jacobs, 2007)
Our papers
are found on the Publications page, with some video abstracts on the Videos page.
Some important collaborators past and present:
- Adnane Achour, Karolinska Institute
- Tim Elliott and Denise Boulanger, Southampton University Medical School
- Peter van Endert, Institut Necker Enfants Malades, Paris
- Hartmut Hengel, Freiburg University Medical School
- Randy Schekman, University of California, Berkeley
- Stefan Stevanović and Hans-Georg Rammensee, Tübingen University
- Mathias Winterhalter, Jacobs University
- Martin Zacharias, Munich Technical University